 Computational Methods
Computational Methods
 Immunology
Immunology
 Alex K. Shalek
Alex K. Shalek
 Bokai Zhu
Bokai Zhu
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Alex K. Shalek
Alex K. Shalek
 Michelle Ramseier
Michelle Ramseier
 Computational Methods
Computational Methods
 Infectious Disease
Infectious Disease
 Medicine
Medicine
 Alex K. Shalek
Alex K. Shalek
 Constantine Tzouanas
Constantine Tzouanas
 Marc Wadsworth II
Marc Wadsworth II
 Computational Methods
Computational Methods
 Infectious Disease
Infectious Disease
 Medicine
Medicine
 Alex K. Shalek
Alex K. Shalek
 Josh Bromley
Josh Bromley
 Marc Wadsworth II
Marc Wadsworth II
 Sarah Nyquist
Sarah Nyquist
 Travis Hughes
Travis Hughes
 Computational Methods
Computational Methods
 Genomics
Genomics
 R&D
R&D
 Statistics
Statistics
 Alex K. Shalek
Alex K. Shalek
 Biology
Biology
 Cancer
Cancer
 Chemistry
Chemistry
 Computational Methods
Computational Methods
 Genomics
Genomics
 R&D
R&D
 Statistics
Statistics
 Technology
Technology
 Alex K. Shalek
Alex K. Shalek
 Ben Mead
Ben Mead
 Conner Kummerlowe
Conner Kummerlowe
 Ivy Liu
Ivy Liu
 Peter Winter
Peter Winter
 Thomas Nok Hin Cheng
Thomas Nok Hin Cheng
 Walaa Kattan
Walaa Kattan
 Biology
Biology
 Cell Atlas
Cell Atlas
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 R&D
R&D
 Alex K. Shalek
Alex K. Shalek
 Biology
Biology
 Cell Atlas
Cell Atlas
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Infectious Disease
Infectious Disease
 R&D
R&D
 Alex K. Shalek
Alex K. Shalek
 James Gatter
James Gatter
 Josh Bromley
Josh Bromley
 Samira Ibrahim
Samira Ibrahim
 Sarah Nyquist
Sarah Nyquist
 Tasneem Jivanjee
Tasneem Jivanjee
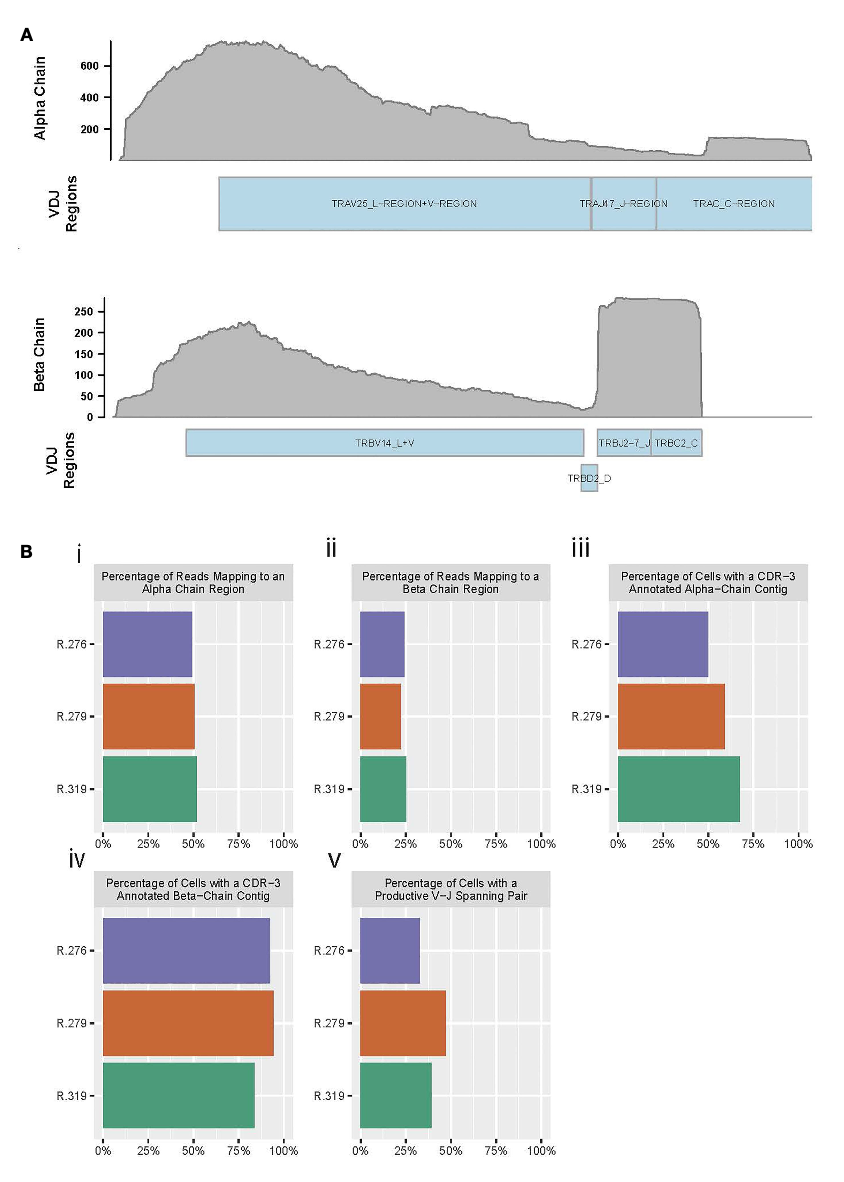
 Biology
Biology
 Cell Atlas
Cell Atlas
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 R&D
R&D
 Technology
Technology
 Alex K. Shalek
Alex K. Shalek
 Jim Kaminski
Jim Kaminski
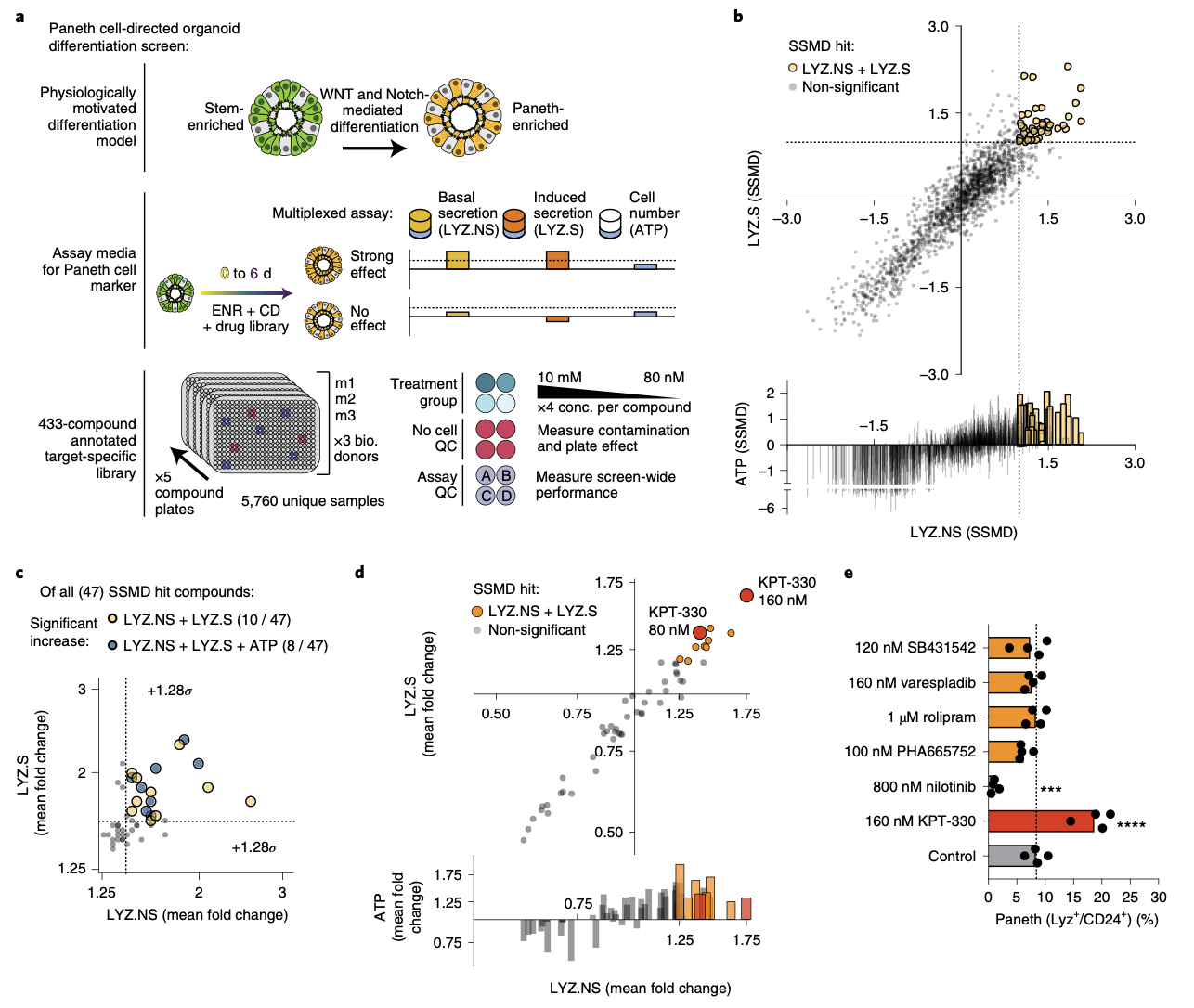
 Biology
Biology
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Medicine
Medicine
 R&D
R&D
 Technology
Technology
 Alex K. Shalek
Alex K. Shalek
 Ben Mead
Ben Mead
 Conner Kummerlowe
Conner Kummerlowe
 José Ordovas-Montañes
José Ordovas-Montañes
 Marko Vukovic
Marko Vukovic
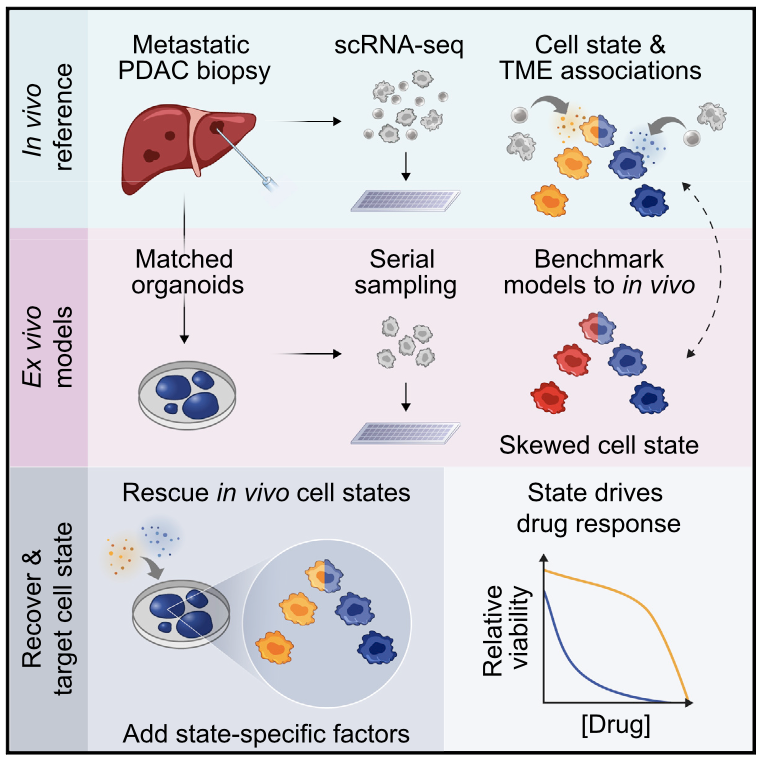
 Cancer
Cancer
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Medicine
Medicine
 R&D
R&D
 Technology
Technology
 Alex K. Shalek
Alex K. Shalek
 Andrew Navia
Andrew Navia
 Jennyfer Galvez-Reyes
Jennyfer Galvez-Reyes
 Nolawit Mulugeta
Nolawit Mulugeta
 Peter Winter
Peter Winter
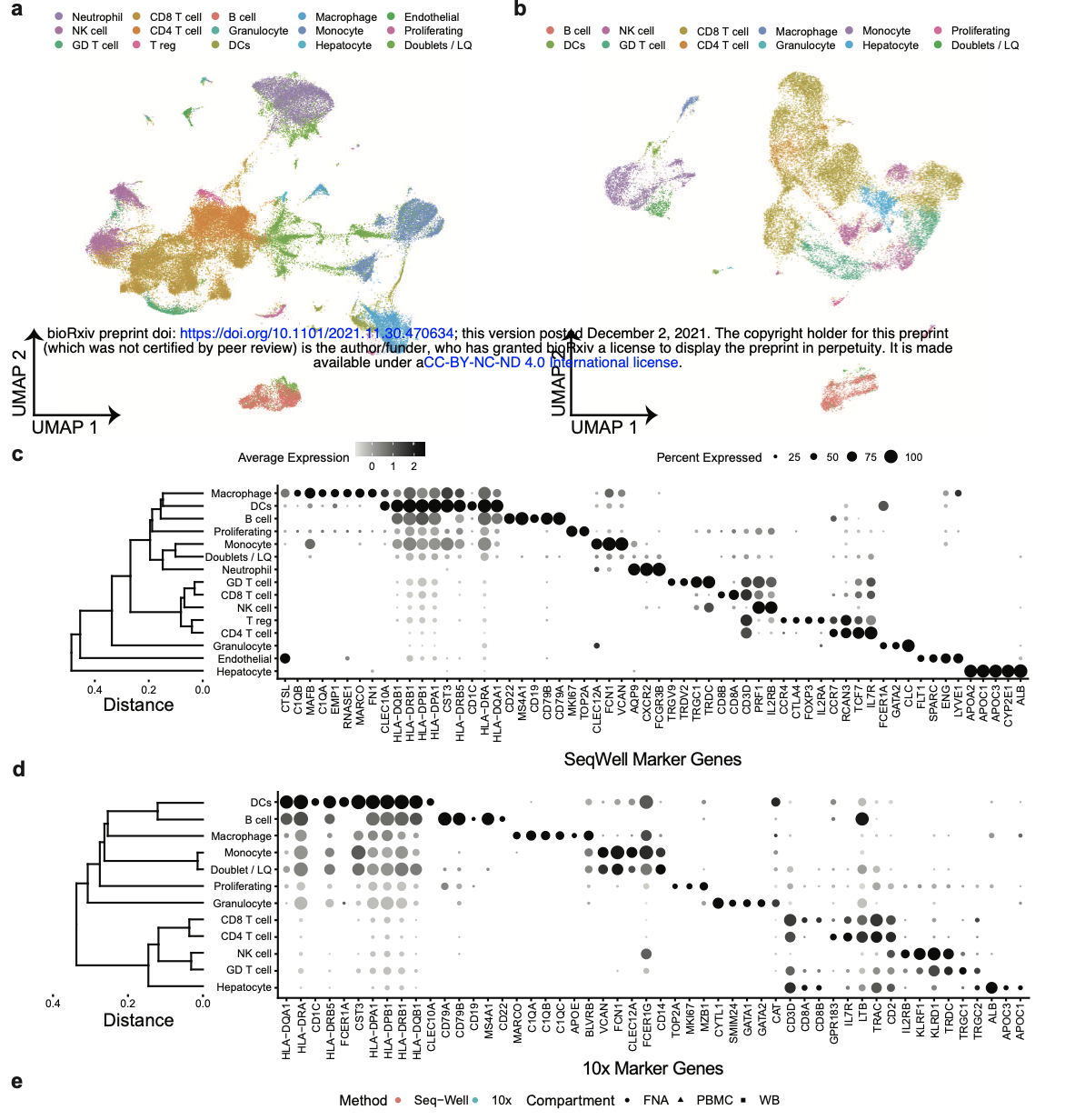
 Biology
Biology
 Cell Atlas
Cell Atlas
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Infectious Disease
Infectious Disease
 R&D
R&D
 Technology
Technology
 Alex Genshaft
Alex Genshaft
 Alex K. Shalek
Alex K. Shalek
 Constantine Tzouanas
Constantine Tzouanas
 Ira Fleming
Ira Fleming
 Jasmin Joseph-Chazan
Jasmin Joseph-Chazan
 Mike Vilme
Mike Vilme
 Nancy Tran
Nancy Tran
 Riley Drake
Riley Drake
 Biology
Biology
 Cell Atlas
Cell Atlas
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Medicine
Medicine
 Alex K. Shalek
Alex K. Shalek
 Benjamin Doran
Benjamin Doran
 José Ordovas-Montañes
José Ordovas-Montañes
 Kyle Kimler
Kyle Kimler
 Biology
Biology
 Cell Atlas
Cell Atlas
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Infectious Disease
Infectious Disease
 Alex K. Shalek
Alex K. Shalek
 Andrew Navia
Andrew Navia
 Carly Ziegler
Carly Ziegler
 Josh Bromley
Josh Bromley
 José Ordovas-Montañes
José Ordovas-Montañes
 Micayla George
Micayla George
 Riley Drake
Riley Drake
 Vincent Miao
Vincent Miao
 Biology
Biology
 Cell Atlas
Cell Atlas
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Infectious Disease
Infectious Disease
 Statistics
Statistics
 Alex K. Shalek
Alex K. Shalek
 Carly Ziegler
Carly Ziegler
 Cell Atlas
Cell Atlas
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Infectious Disease
Infectious Disease
 Statistics
Statistics
 Alex K. Shalek
Alex K. Shalek
 Carly Ziegler
Carly Ziegler
 Marc Wadsworth II
Marc Wadsworth II
 Sam Allon
Sam Allon
 Sarah Nyquist
Sarah Nyquist
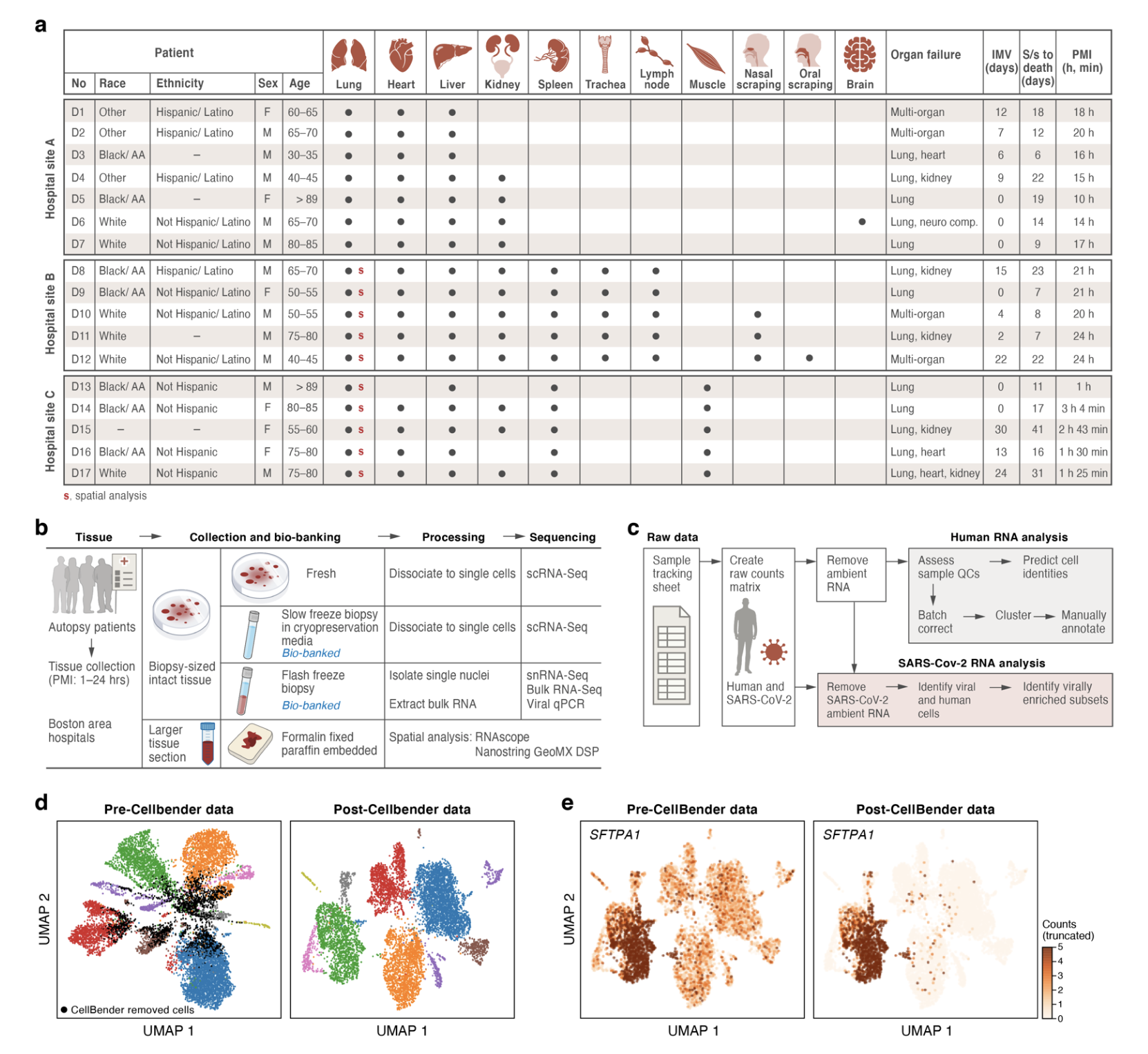
 Biology
Biology
 Cell Atlas
Cell Atlas
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Infectious Disease
Infectious Disease
 Statistics
Statistics
 Alex K. Shalek
Alex K. Shalek
 Carly Ziegler
Carly Ziegler
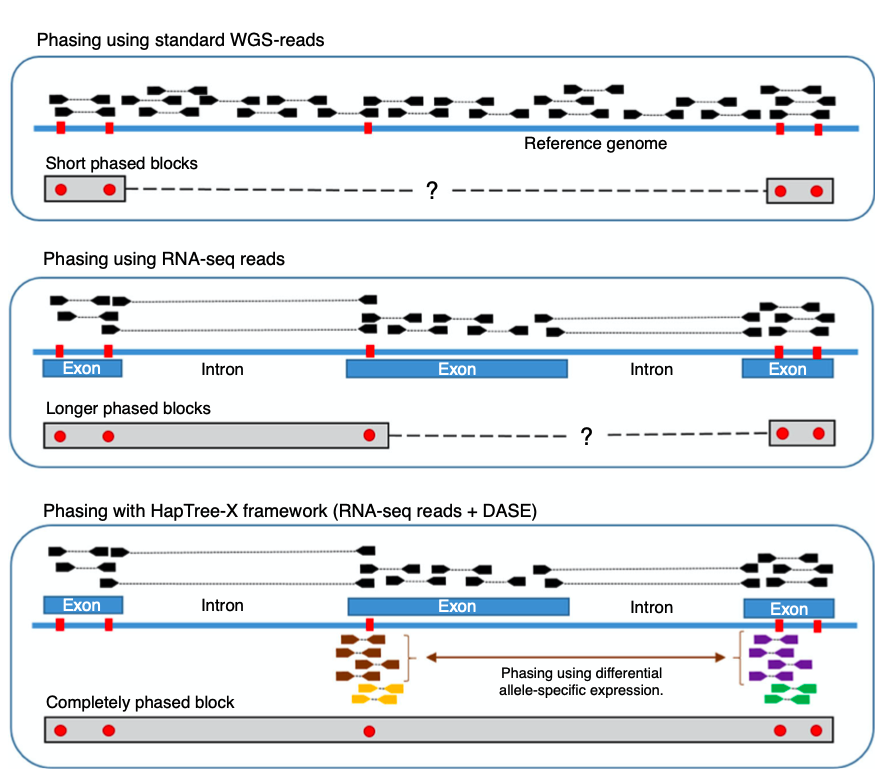
 Biology
Biology
 Computational Methods
Computational Methods
 Statistics
Statistics
 Alex K. Shalek
Alex K. Shalek
 Sarah Nyquist
Sarah Nyquist
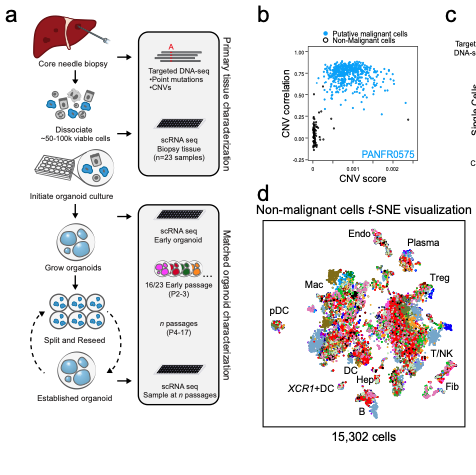
 Biology
Biology
 Cancer
Cancer
 Cell Atlas
Cell Atlas
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Medicine
Medicine
 Technology
Technology
 Alex K. Shalek
Alex K. Shalek
 Andrew Navia
Andrew Navia
 Jennyfer Galvez-Reyes
Jennyfer Galvez-Reyes
 Nolawit Mulugeta
Nolawit Mulugeta
 Peter Winter
Peter Winter
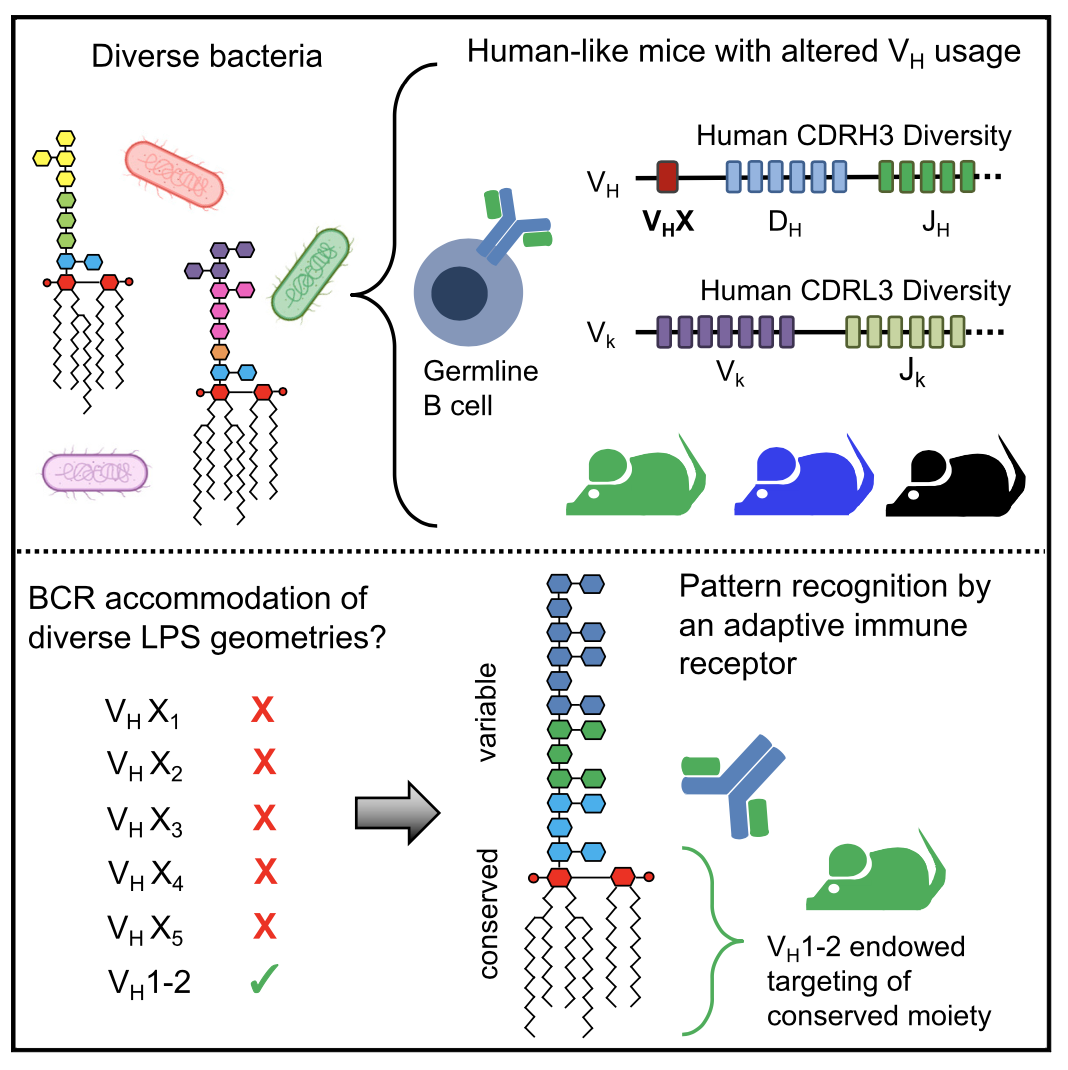
 Biology
Biology
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Infectious Disease
Infectious Disease
 Alex K. Shalek
Alex K. Shalek
 James Gatter
James Gatter
 Sam Kazer
Sam Kazer
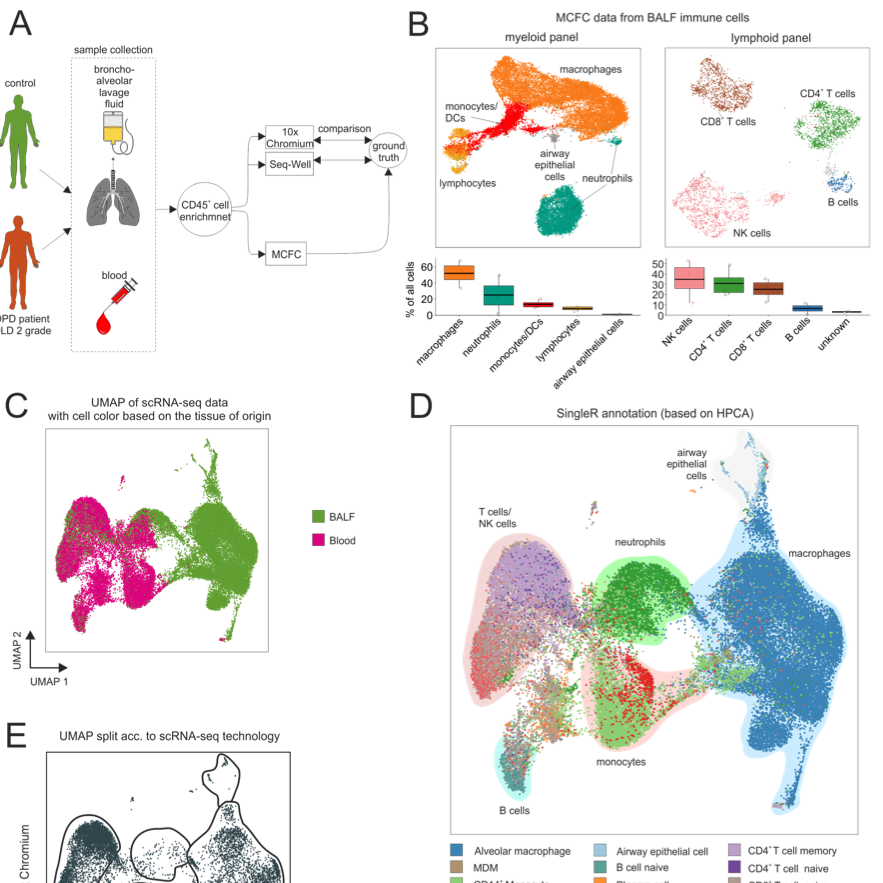
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Medicine
Medicine
 Alex K. Shalek
Alex K. Shalek
 Marc Wadsworth II
Marc Wadsworth II
 Travis Hughes
Travis Hughes
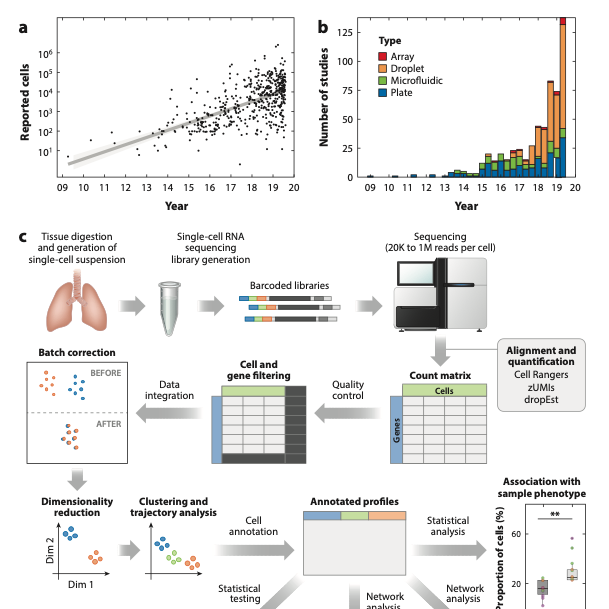
 Biology
Biology
 Computational Methods
Computational Methods
 Genomics
Genomics
 Alex K. Shalek
Alex K. Shalek
 Sarah Nyquist
Sarah Nyquist
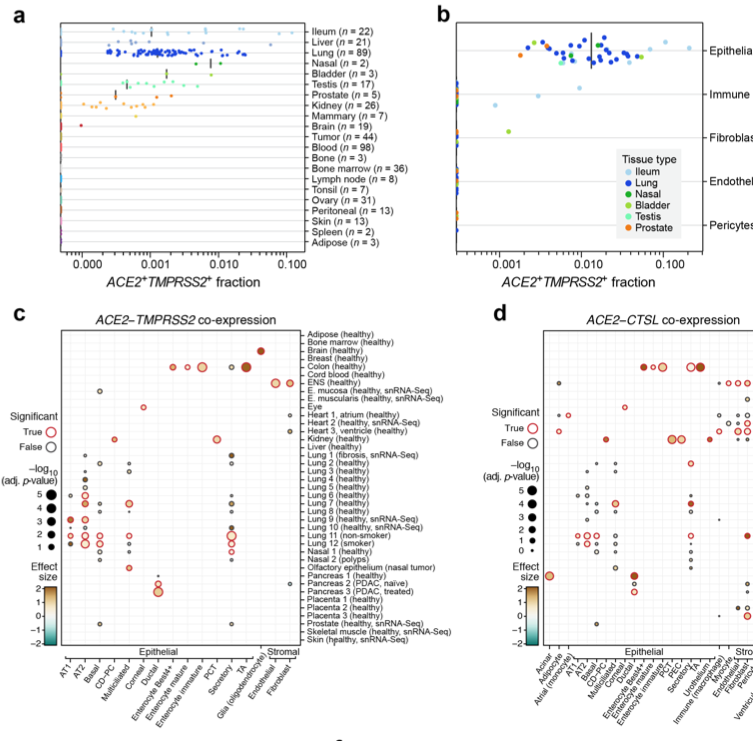
 Biology
Biology
 Cell Atlas
Cell Atlas
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Infectious Disease
Infectious Disease
 Statistics
Statistics
 Alex K. Shalek
Alex K. Shalek
 Carly Ziegler
Carly Ziegler
 José Ordovas-Montañes
José Ordovas-Montañes
 Marc Wadsworth II
Marc Wadsworth II
 Sam Allon
Sam Allon
 Sarah Nyquist
Sarah Nyquist
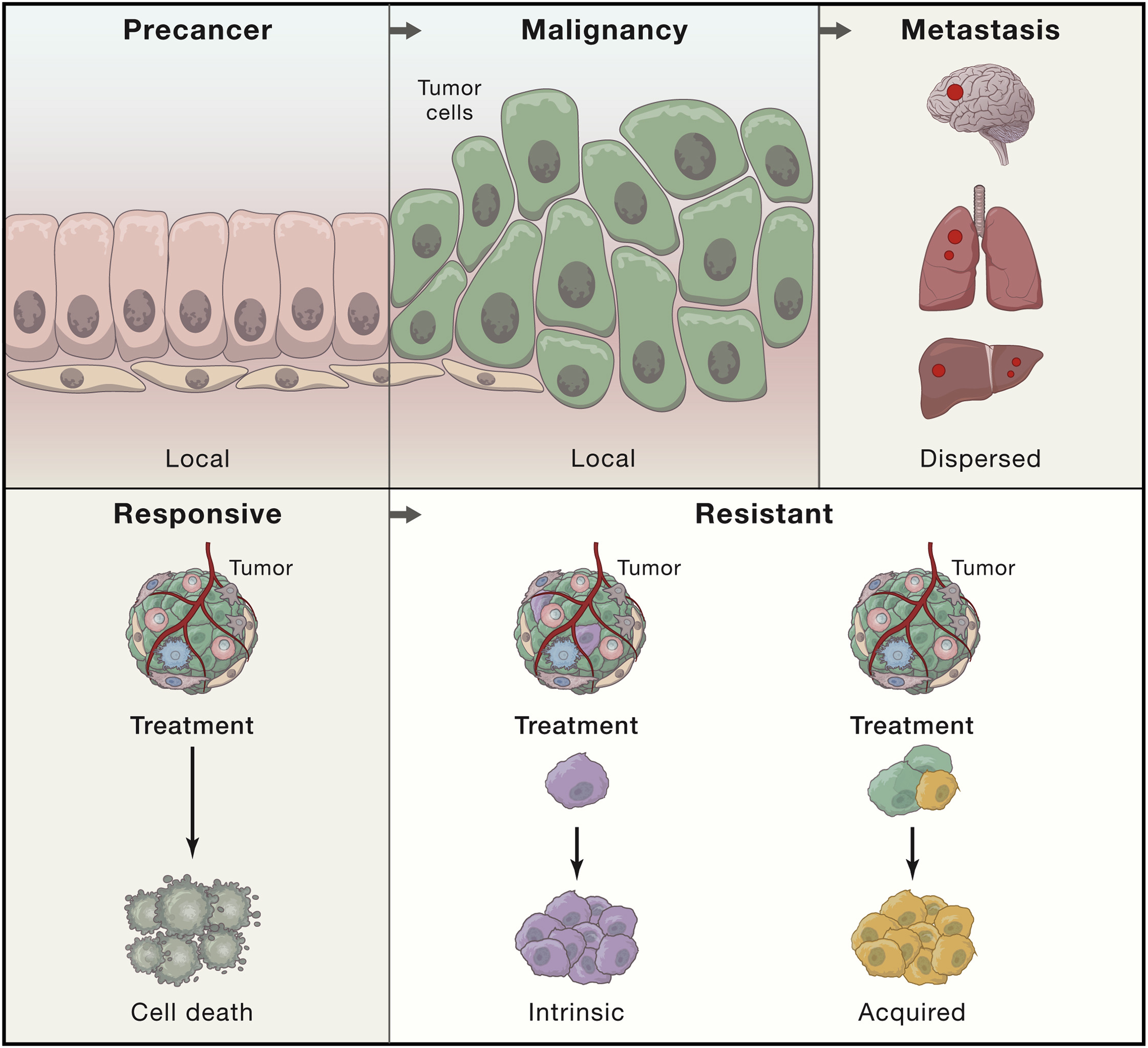
 Biology
Biology
 Cancer
Cancer
 Cell Atlas
Cell Atlas
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Medicine
Medicine
 Alex K. Shalek
Alex K. Shalek
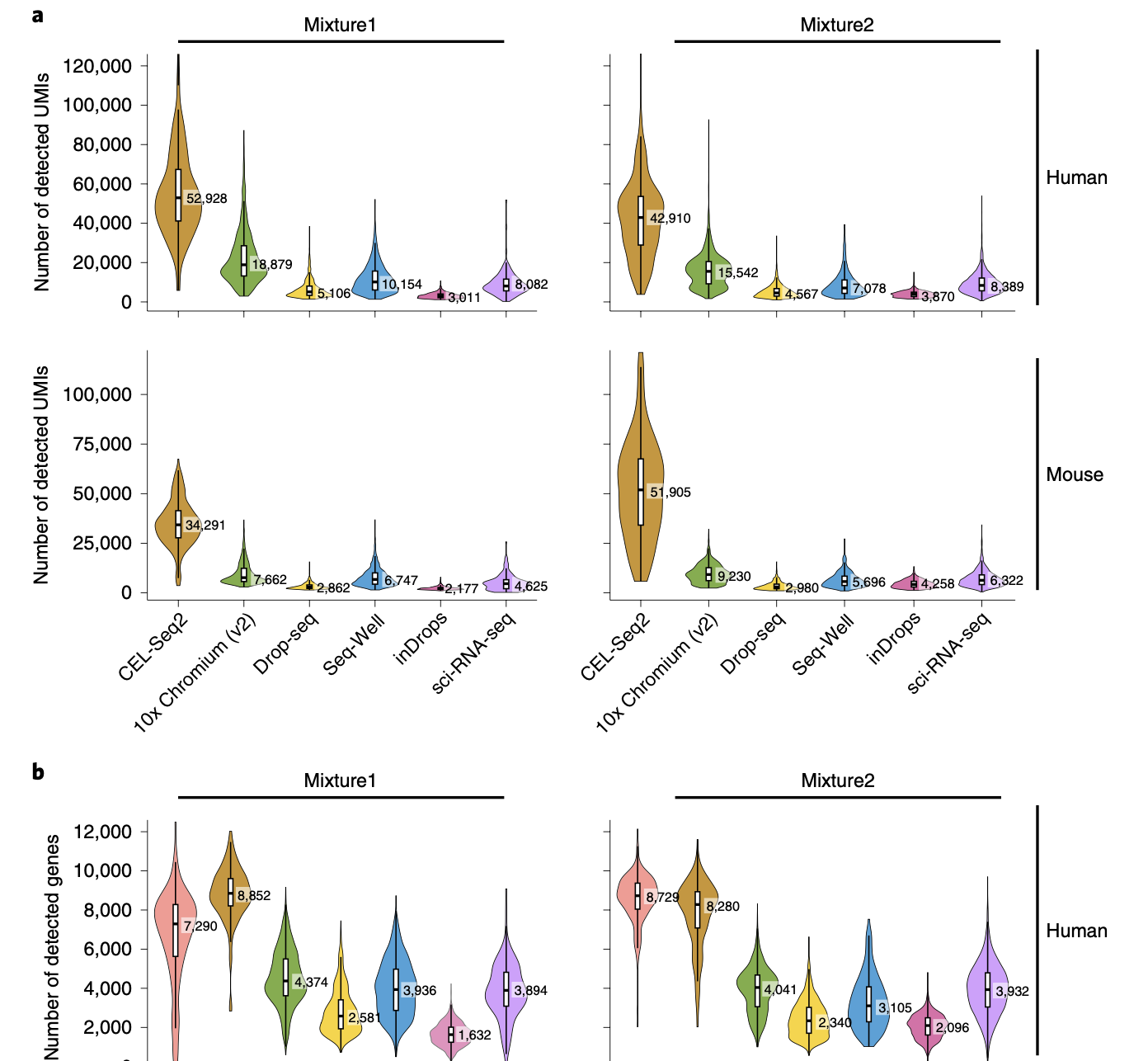
 Computational Methods
Computational Methods
 Genomics
Genomics
 R&D
R&D
 Technology
Technology
 Alex K. Shalek
Alex K. Shalek
 Marc Wadsworth II
Marc Wadsworth II
 Shaina Carroll
Shaina Carroll
 Travis Hughes
Travis Hughes

 Biology
Biology
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Infectious Disease
Infectious Disease
 Statistics
Statistics
 Alex K. Shalek
Alex K. Shalek
 Carly Ziegler
Carly Ziegler
 José Ordovas-Montañes
José Ordovas-Montañes
 Sam Kazer
Sam Kazer
 Sarah Nyquist
Sarah Nyquist
 Shaina Carroll
Shaina Carroll
 Toby Aicher
Toby Aicher
 Vincent Miao
Vincent Miao

 Biology
Biology
 Cell Atlas
Cell Atlas
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Infectious Disease
Infectious Disease
 Medicine
Medicine
 Benjamin Doran
Benjamin Doran
 Carly Ziegler
Carly Ziegler
 Constantine Tzouanas
Constantine Tzouanas
 James Gatter
James Gatter
 Marc Wadsworth II
Marc Wadsworth II
 Marko Vukovic
Marko Vukovic
 Sam Allon
Sam Allon
 Sarah Nyquist
Sarah Nyquist
 Shaina Carroll
Shaina Carroll
 Vincent Miao
Vincent Miao
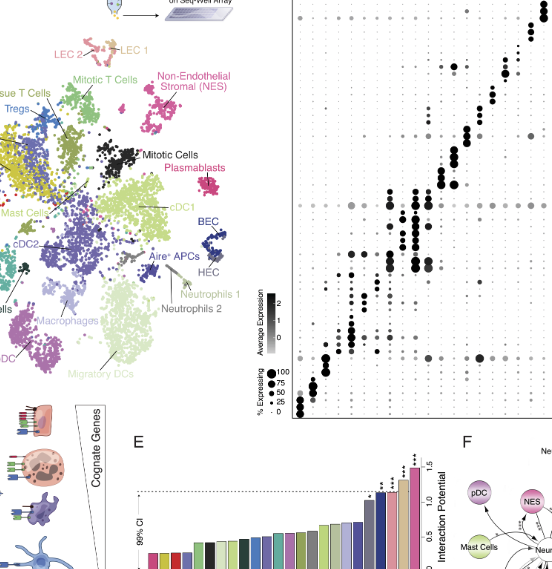
 Biology
Biology
 Cell Atlas
Cell Atlas
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Alex K. Shalek
Alex K. Shalek
 Carly Ziegler
Carly Ziegler
 José Ordovas-Montañes
José Ordovas-Montañes
 Marko Vukovic
Marko Vukovic

 Computational Methods
Computational Methods
 Genomics
Genomics
 R&D
R&D
 Technology
Technology
 Alex K. Shalek
Alex K. Shalek
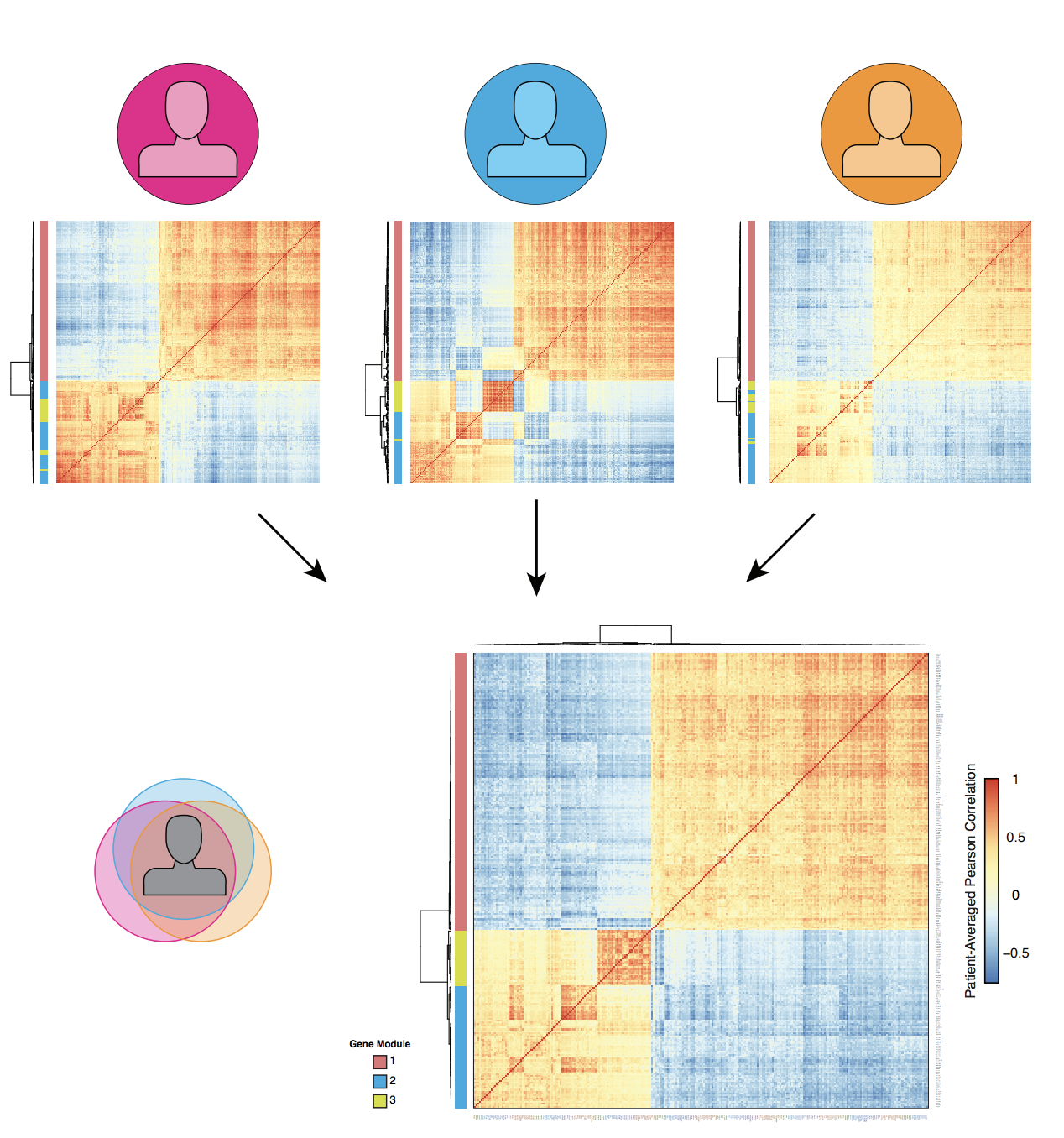
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Infectious Disease
Infectious Disease
 Medicine
Medicine
 Alex K. Shalek
Alex K. Shalek
 José Ordovas-Montañes
José Ordovas-Montañes
 Kellie Kolb
Kellie Kolb

 Computational Methods
Computational Methods
 Statistics
Statistics
 Alex K. Shalek
Alex K. Shalek
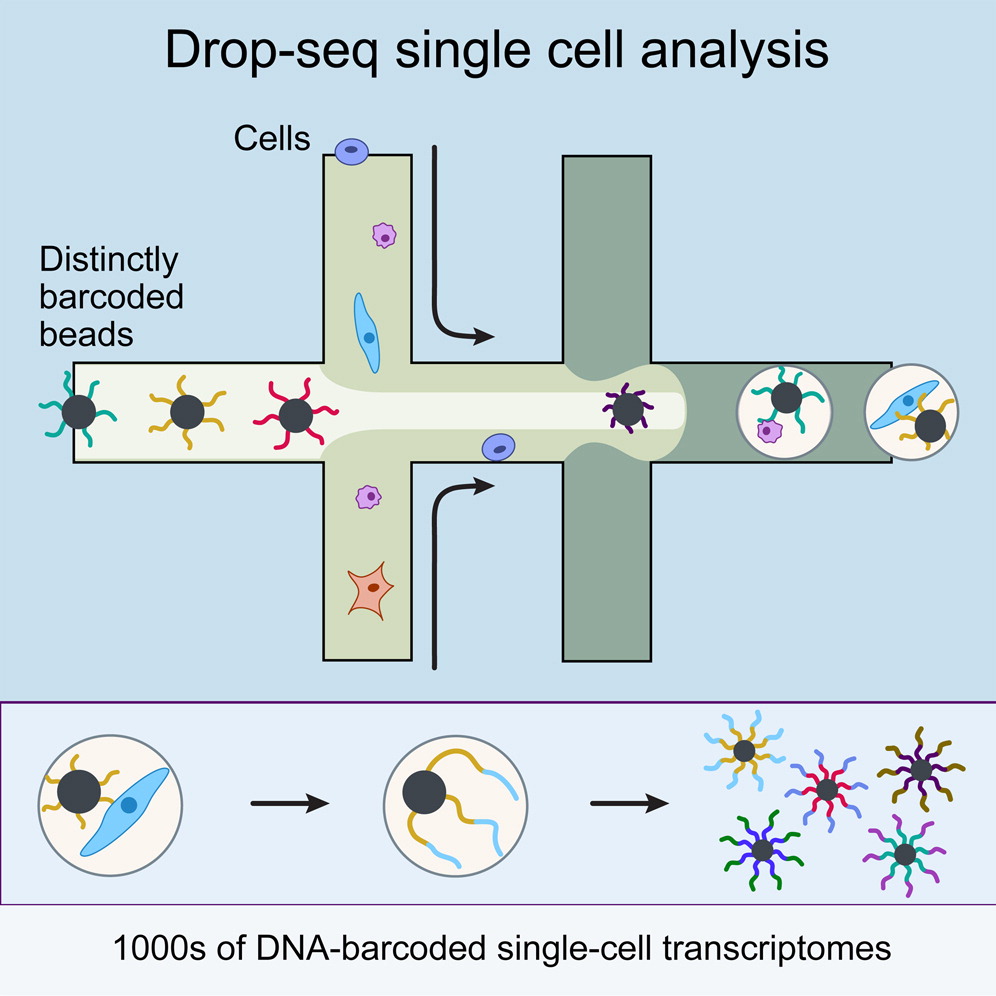
 Genomics
Genomics
 R&D
R&D
 Technology
Technology
 Alex K. Shalek
Alex K. Shalek