Our biological applications focus on how immune cells interface with diverse, dynamic, tissue-specific cell types/states and signals to maintain tolerance and defend against pathogens and malignancies. Contrasting cellular ecosystems in health and disease (e.g., inflammation, infection, or cancer) with collaborators around the globe, we identify putative intra- and intercellular mechanisms for maintaining biological function under stress and aberrant behaviors associated with pathology. By methodically testing and manipulating features linked to enhanced tissue function (e.g., control of HIV-1) or dysfunction (e.g., polyposis in allergic inflammation), we work to achieve an actionable understanding of the intra- and extracellular circuits that underpin tissue-level behaviors toward helping realize rational strategies to improve human health.
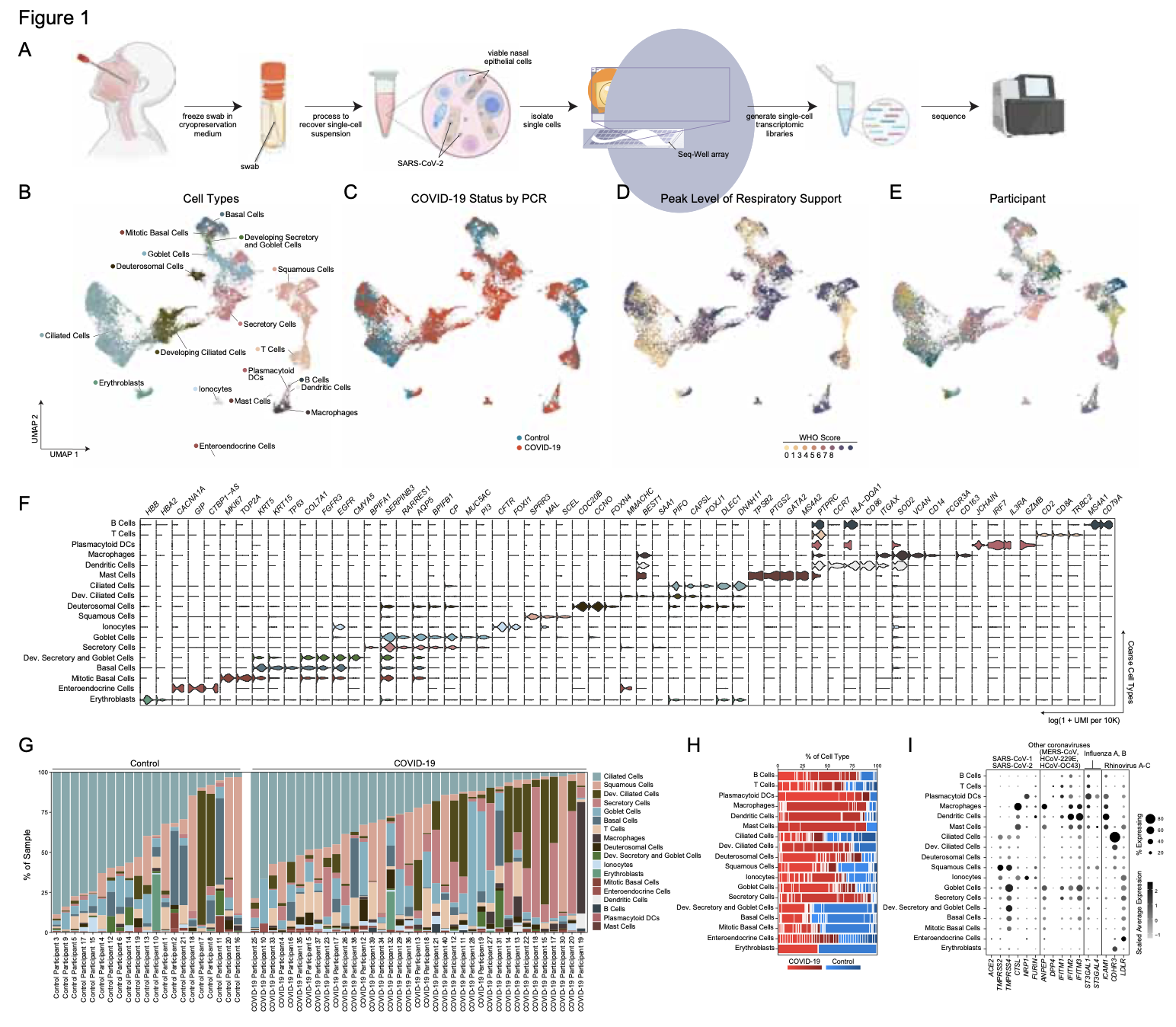
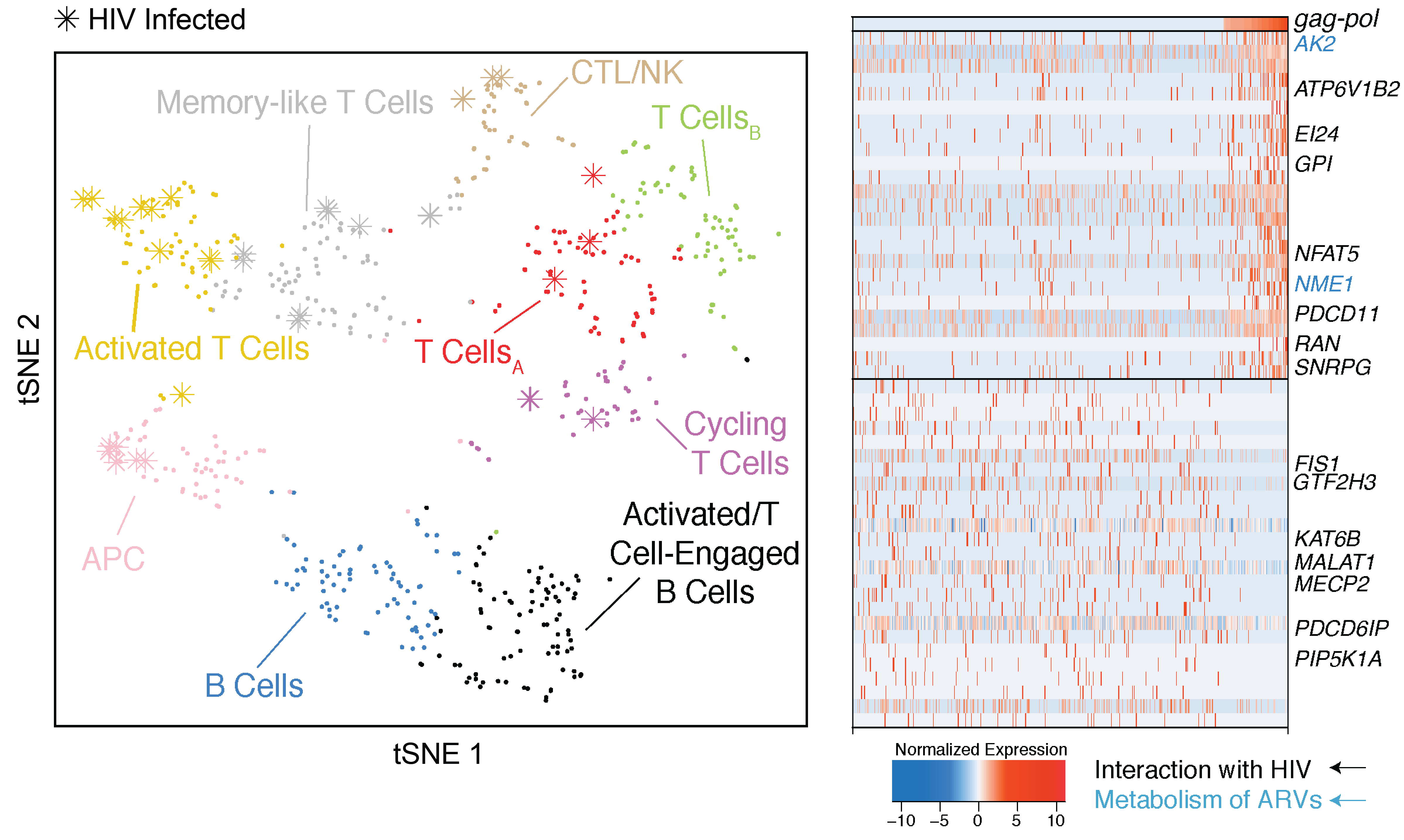

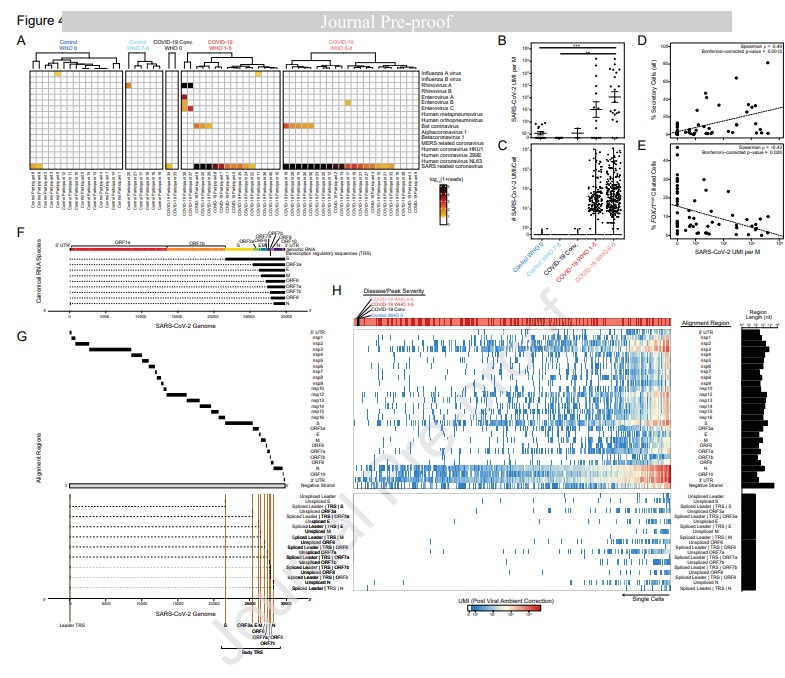
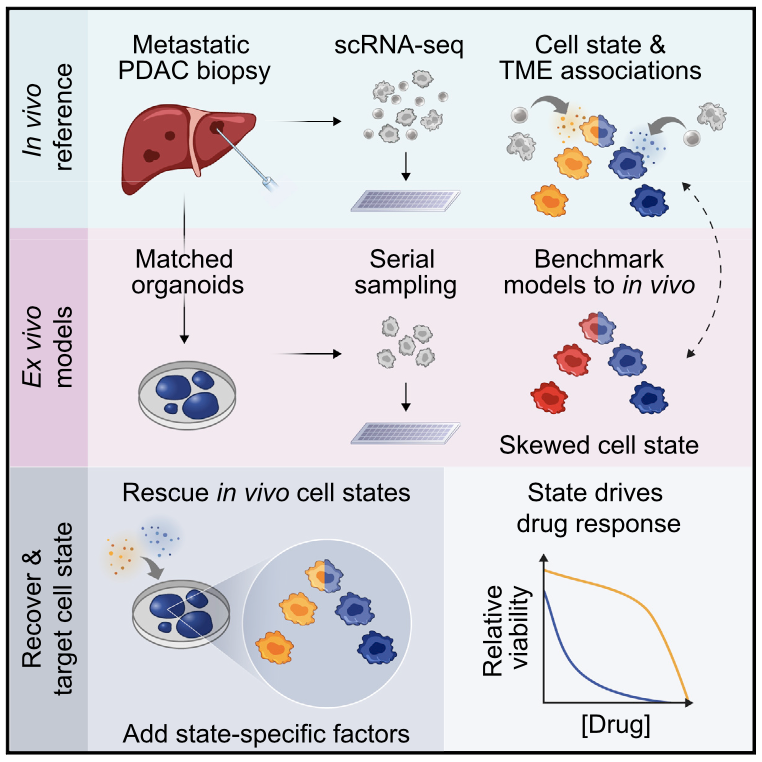
We lack effective treatments and preventions for many of the most challenging infectious diseases, many of which disproportionately impact those in low- and middle-income countries or traditionally marginalized communities.
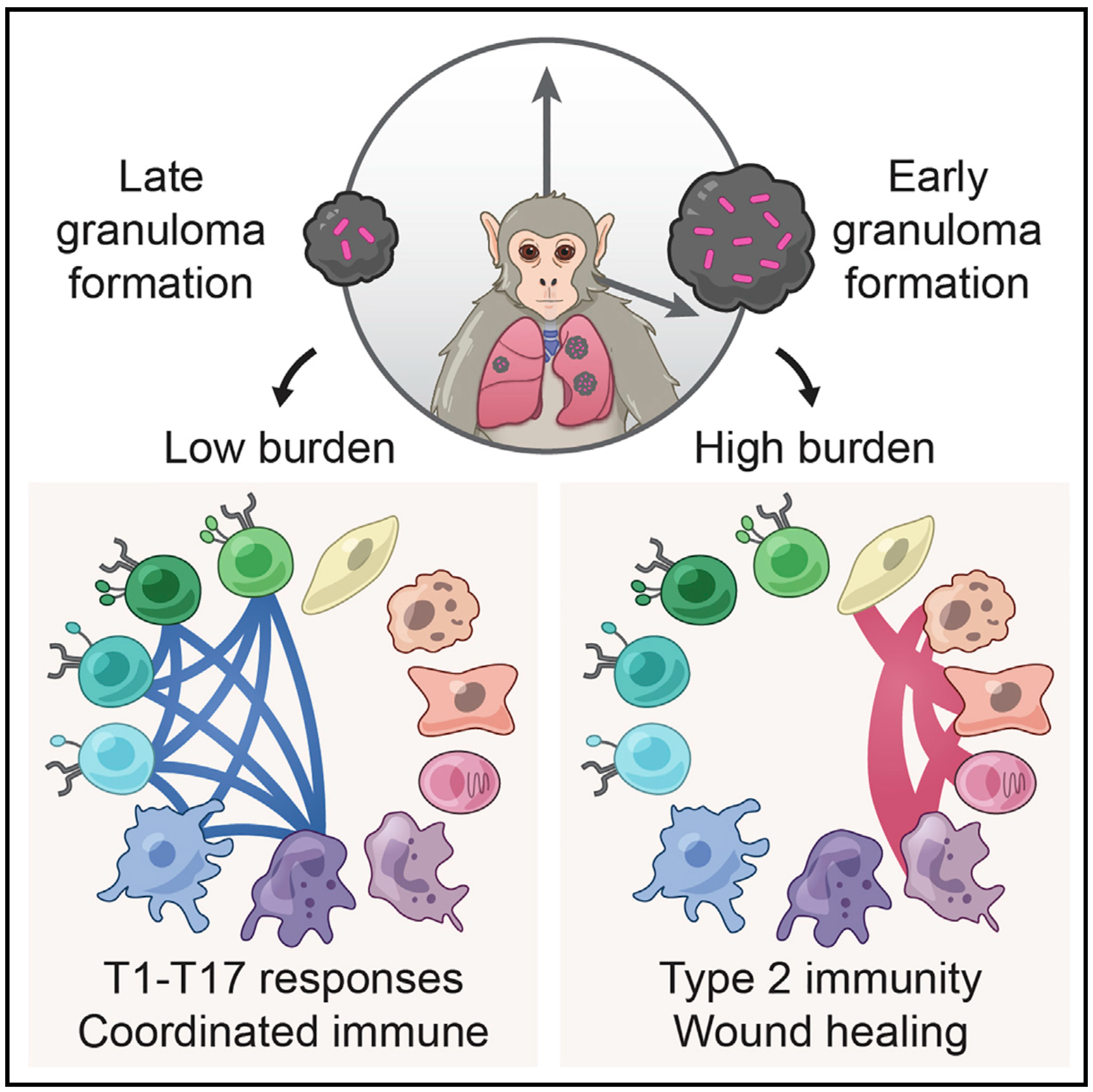
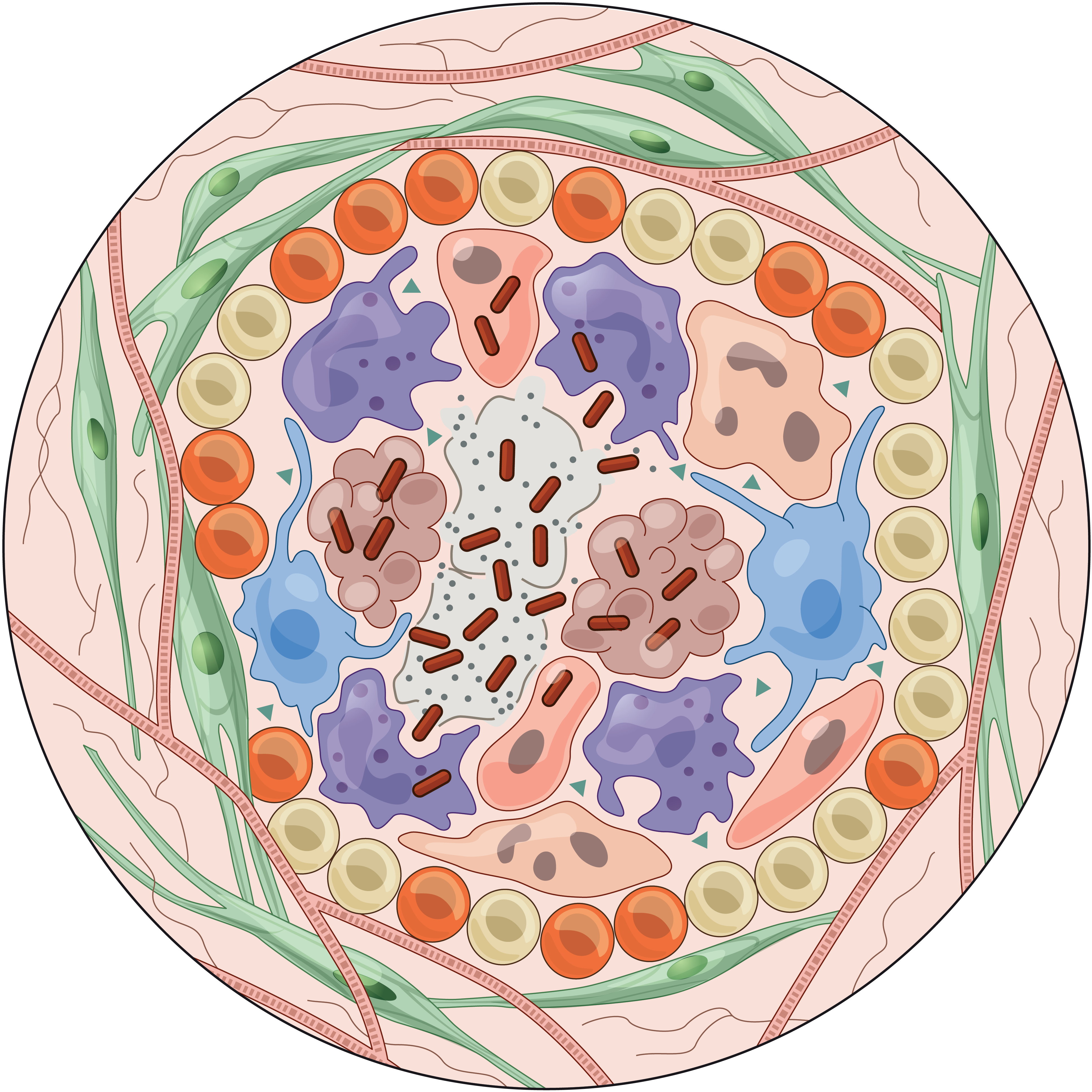
To help address this, we have established and enabled multi-group, multi-country partnerships to deploy and adapt cutting-edge genomic tools. By examining how cells dynamically alter their states, individually and collectively, during disease and/or its resolution in acute and chronic infections—e.g., tuberculosis, HIV/SHIV, hepatitis, malaria, leprosy, flu, SARS-CoV-2, and ebola—we have uncovered cellular and molecular features of pathogen control or pathology to potentiate or counteract, respectively. Illustratively, in tuberculosis, we identified a functional role for cytotoxic CD8 and hybrid type1-type17 T cells in control of infection in the lung and links between mast, plasma, and endothelial cell abundance (type-2 immune responses) and bacterial burden. We have also built methods for examining pathogens within individual host cells to define their dynamic interdependence and identify potentially restrictive host factors.
We are currently working to identify the drivers of common host responses to distinct perturbations and their targetability, as well as the impact of different interventions (e.g., vaccines).
Representative Publication: Ziegler et al., Cell, 2021
Representative Publication: Gideon et al., Immunity, 2022
Representative Publication: Kazer et al., Nature Medicine, 2020
 Aleth Gaillard
Aleth Gaillard
 Carly Ziegler
Carly Ziegler
 Conner Kummerlowe
Conner Kummerlowe
 Constantine Tzouanas
Constantine Tzouanas
 Josh Bromley
Josh Bromley
 Marc Wadsworth II
Marc Wadsworth II
 Riley Drake
Riley Drake
 Samira Ibrahim
Samira Ibrahim
 Sarah Nyquist
Sarah Nyquist
 Vincent Miao
Vincent Miao
 Biology
Biology
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Infectious Disease
Infectious Disease
 Medicine
Medicine
 Microbiology
Microbiology
 R&D
R&D
 Statistics
Statistics
 Technology
Technology
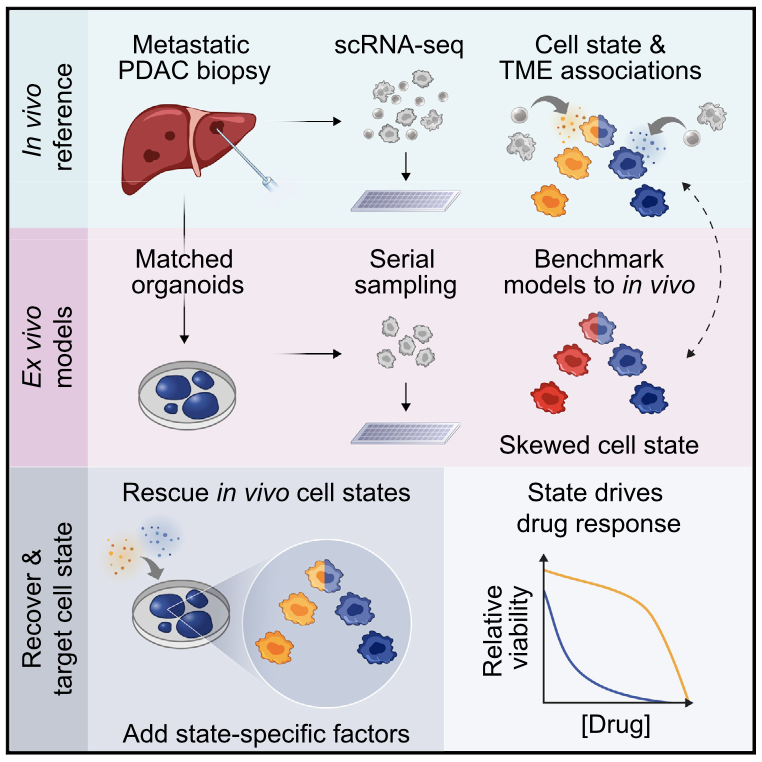
Immune responses play a critical role in preventing tumorigenesis. Sometimes, however, they are ineffectual and can even drive/support malignancy.
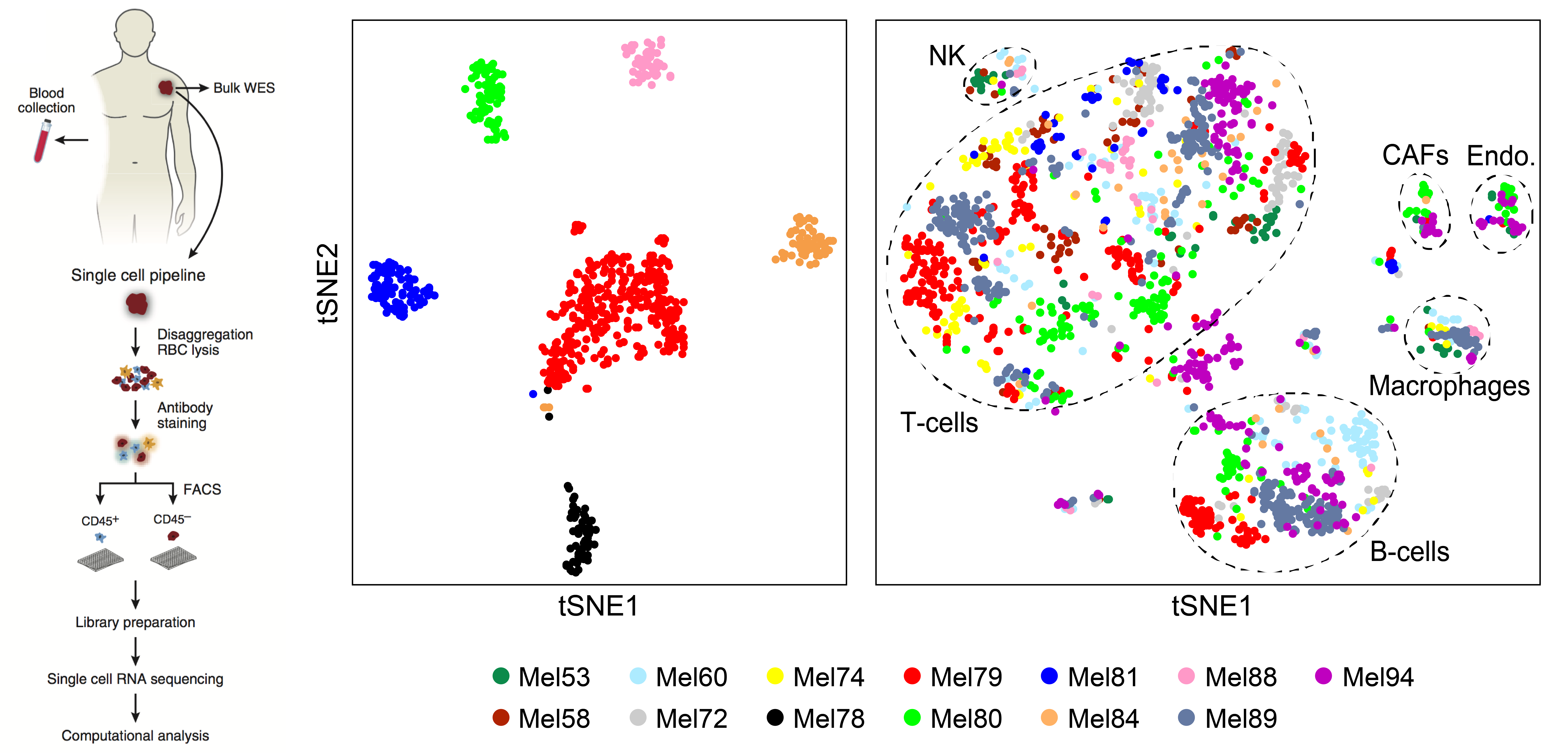
We have examined how cancer cells alter and are influenced by their tumor microenvironments (TMEs), and the impact this has on therapeutic responses. Illustratively, in Pancreatic Ductal Adenocarcinoma (PDAC), by profiling liver metastases and matched organoid models, we showed: 1. associations between TME and malignant cell state composition; 2. that autocrine and paracrine signaling can drive malignant cell state transitions, even in an isogenic background, altering the efficacy of frontline chemotherapies; and, 3. that microenvironmental manipulations can be used to control malignant state, and thereby drug responses, rationally, and to improve model fidelity for screening potential therapies. This and related work highlight the potential utility of modulating indirect target cells (T cells in the PDAC TME or basal cells in allergic inflammation) to enhance cures and preventions.
We are now systematically expanding this work to define how additional environmental and cell-intrinsic factors influence malignant cell state plasticity in PDAC and other cancers toward enhancing treatments.
Representative Publication: Tirosh et al., Science, 2016
Representative Publication: Raghavan et al., Cell, 2021
Representative Publication: Prakadan et al., Nature Communications, 2021
 Alex Genshaft
Alex Genshaft
 Andrew Navia
Andrew Navia
 Carly Ziegler
Carly Ziegler
 Jay Prakadan
Jay Prakadan
 Jennyfer Galvez-Reyes
Jennyfer Galvez-Reyes
 Kellie Kolb
Kellie Kolb
 Marc Wadsworth II
Marc Wadsworth II
 Nolawit Mulugeta
Nolawit Mulugeta
 Sam Allon
Sam Allon
 Sarah Nyquist
Sarah Nyquist
 Biology
Biology
 Cancer
Cancer
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Medicine
Medicine
 Microbiology
Microbiology
 R&D
R&D
 Statistics
Statistics
 Technology
Technology
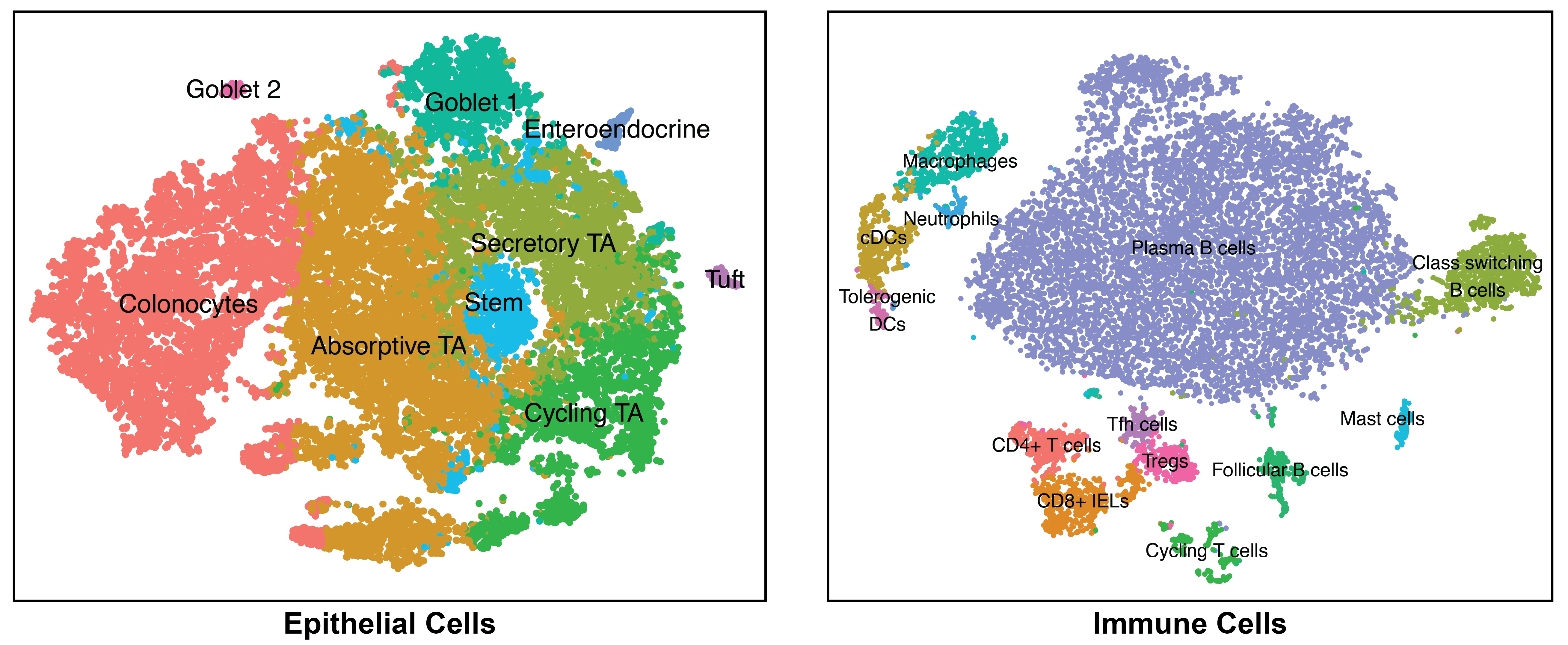
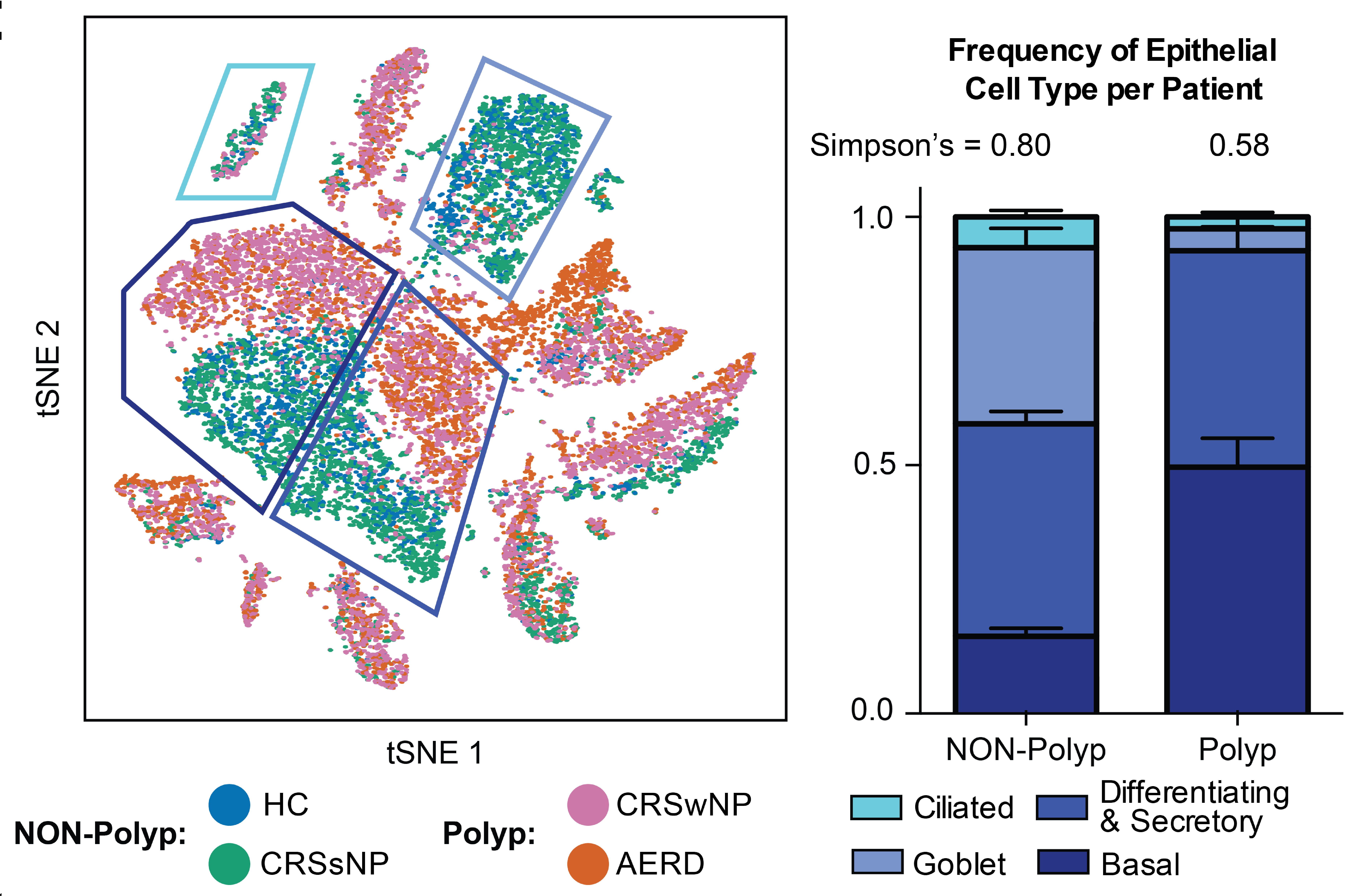
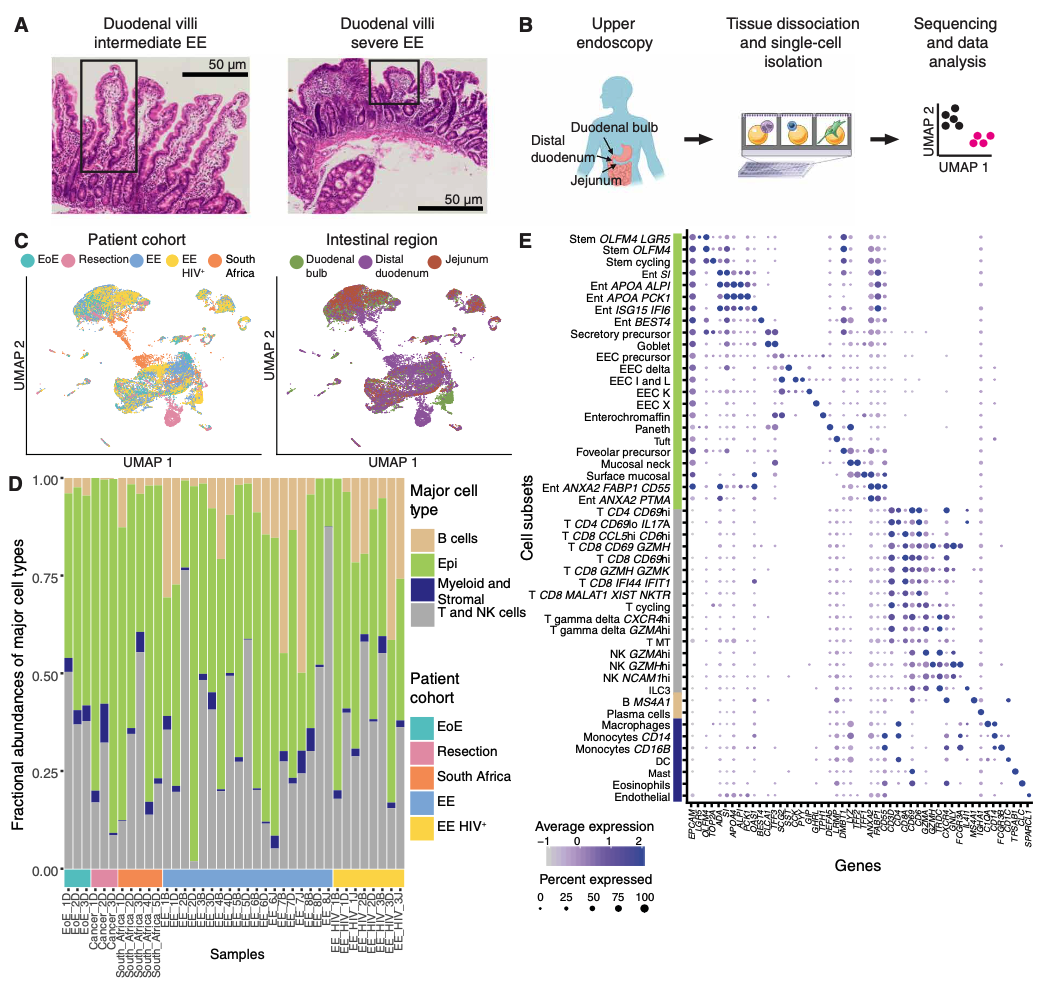
We are exposed to a constant flux of external biochemical and physical stimuli as we age. Despite variability in our overall experiences and exact constitutions, our individual tissues typically manage to maintain functionality, though each can differ in its resilience to distinct stressors.
We have characterized how differences in cellular composition and communication impact tissue fitness and have identified responses and subsequent adaptations that drive chronic dysfunction. For example, although aberrant immune activity can precipitate allergic inflammatory diseases, therapies targeting immune cells and signaling are only successful in some, suggesting chronicity may involve alternative mechanisms. Previously, we helped demonstrate that dysregulated type-2 immune signaling, driven by environmental allergens, can impact tissue health in the upper airway through generating dysfunctional basal epithelial stem cells. These stem cells can then contribute to persistence by serving as repositories for allergic inflammatory memories, altering the integrity and functional output of the nasal epithelium. Our work, with that of others, suggests generalizable principles for cellular memory, and informs where and how tissues should be targeted to support health or restore function. We have since further investigated how tissue-resident cellular subsets participate in, and are shaped by, environmental exposures at barrier tissues and the functional consequences of these experiences.
We are now working to develop a more holistic appreciation for how different intra- and extracellular factors (e.g., genetics and integrated exposure history, respectively) influence barrier tissue function.
Representative Publication: Ordovas-Montanes et al., Nature, 2018
Representative Publication: Kummerlowe et al., Science Translational Medicine, 2022
Representative Publication: Ziegler et al., bioRxiv, 2021
 Benjamin Doran
Benjamin Doran
 Conner Kummerlowe
Conner Kummerlowe
 Ira Fleming
Ira Fleming
 Josh Bromley
Josh Bromley
 Marc Wadsworth II
Marc Wadsworth II
 Marko Vukovic
Marko Vukovic
 Nolawit Mulugeta
Nolawit Mulugeta
 Sam Allon
Sam Allon
 Sarah Nyquist
Sarah Nyquist
 Vincent Miao
Vincent Miao
 Biology
Biology
 Cell Atlas
Cell Atlas
 Chemistry
Chemistry
 Computational Methods
Computational Methods
 Genomics
Genomics
 Immunology
Immunology
 Medicine
Medicine
 R&D
R&D
 Statistics
Statistics
 Technology
Technology