

Computational cohort materials from March 2023 Ghana training...

The Alexandria project aims to facilitate single-cell data analysis for users through providing an accessible data repository along with workflows, notebooks, and scripts...
The Shalek Lab is committed to open science and shares code and protocols on GitHub....
Protocol for integrating CITE-seq with well-based scRNA-seq protocols....
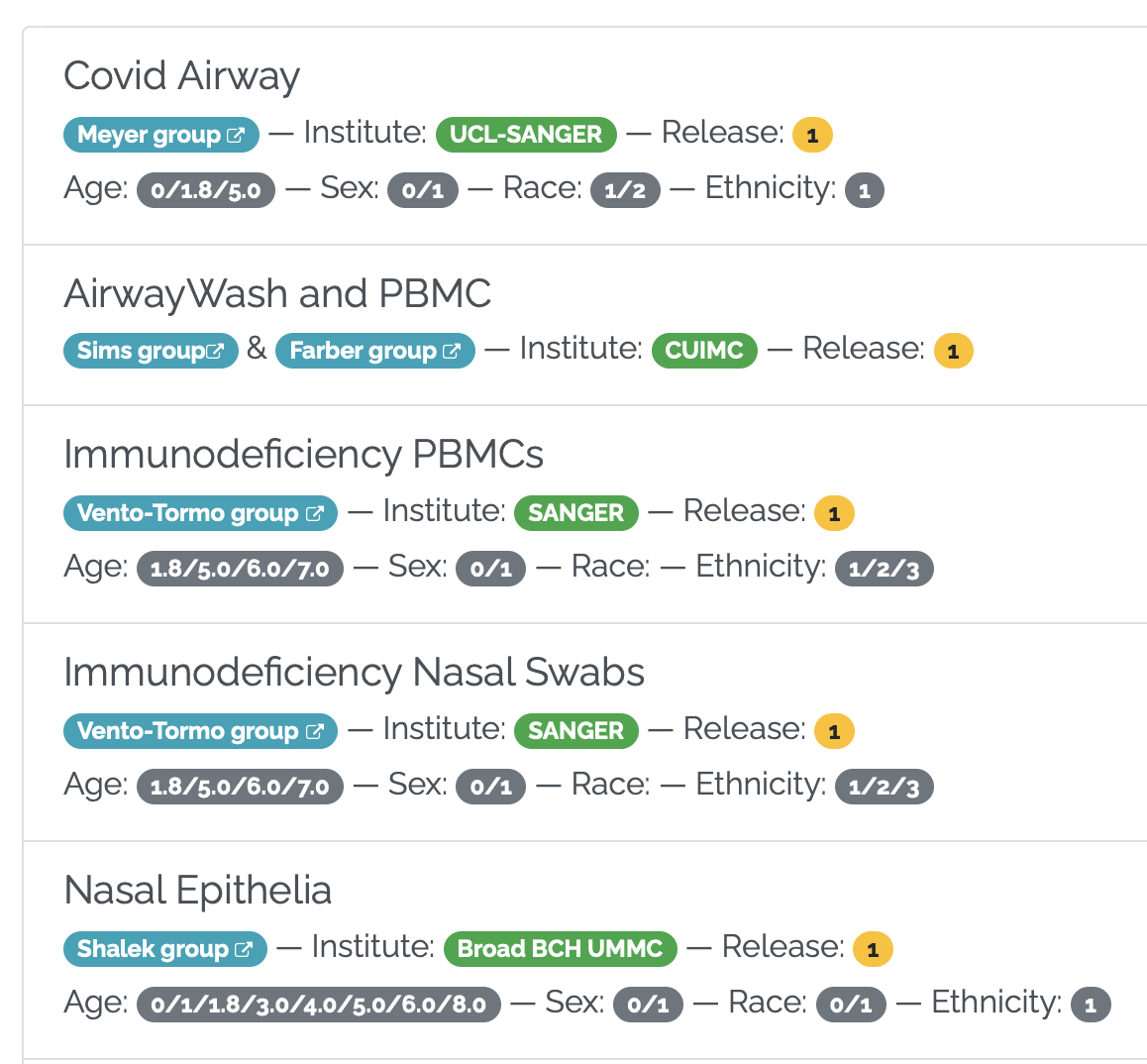
Our nasal epithelial COVID-19 dataset, along with COVID-19 datasets from other genomics groups, can now be found at covid19cellatlas.org. This work was sponsored by the Chan-Zuckerberg Initiative....

The following resources have been compiled from various sources. We would like to thank the authors of each for their work. The Shalek Lab takes no credit for any of the information provided via these links. 26 Ways to be in the Struggle Anti-racist Resources for White People Letters for Black Lives Revolutionary Left Radio: […]...

Overview In light of the global effort to better understand the new SARS-CoV-2 virus, we and other researchers from the HCA Lung Biological Network and beyond have begun an initiative to investigate datasets from relevant tissues profiled as part of other ongoing studies. These studies represent, for example, large efforts to characterize HIV, Mtb, and […]...

The full expression matrix and associated metadata from the publication “Integrated Single-Cell Analysis of Multicellular Immune Dynamics during Hyperacute HIV-1 Infection” can be downloaded from the Alexandria Project hosted through the Single Cell Portal at the Broad Institute. Please visit the interactive dataset here to access and explore the data online. See below for an extended Supplementary […]...
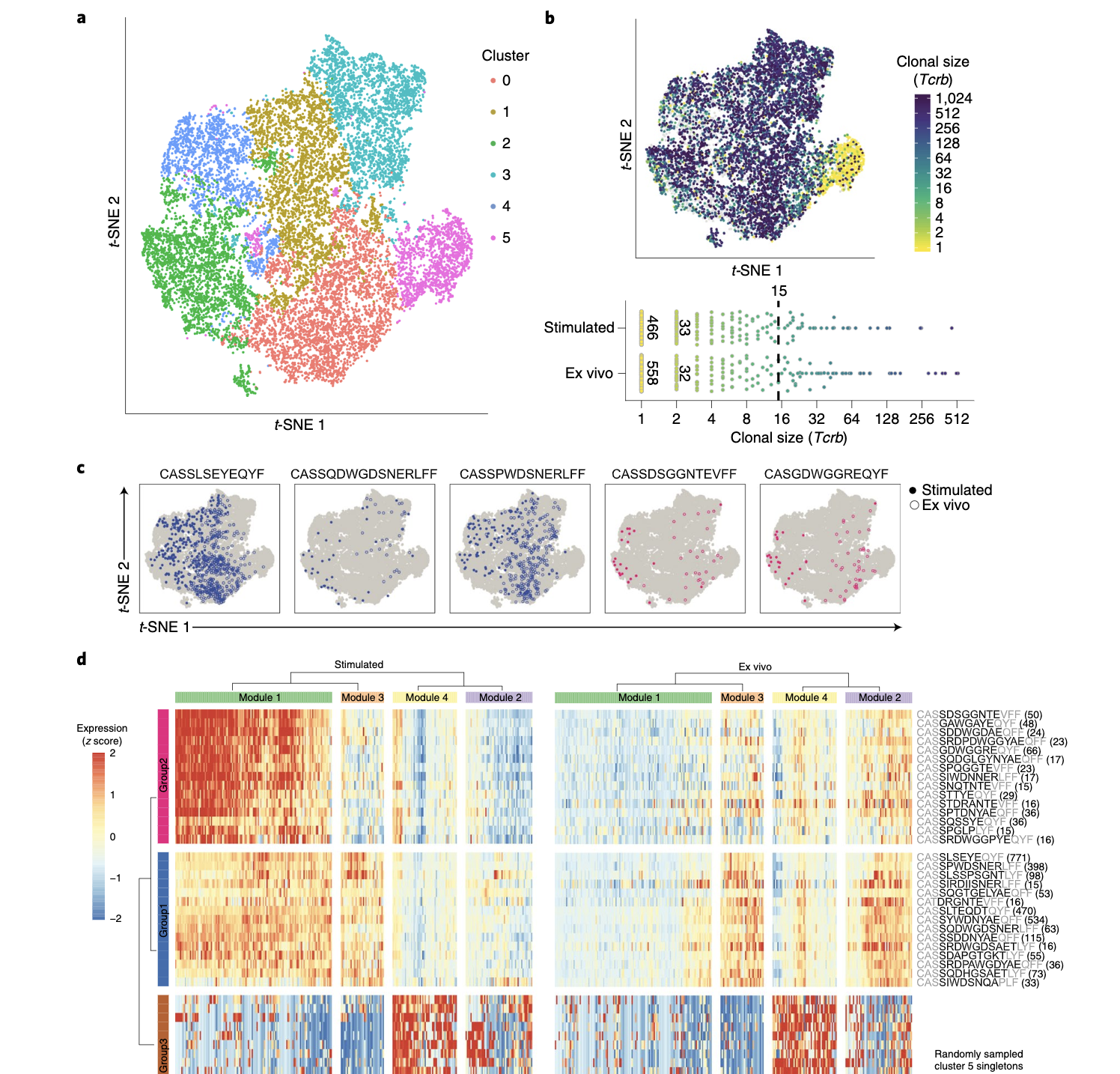
By affinity capture and amplification of TCR transcripts from whole-transcriptome libraries, TCR CDR3 sequences can be recovered from 3′-barcoded scRNA-seq libraries (e.g. Seq-Well, Drop-seq, etc.). This method can be applied post-hoc, allowing for the capture of additional information from archived samples. The protocol can also be found here....
Download Raw Data Download Metadata scRNA-seq data captured on Seq-Well for the core 18,036 cell data set as clustered and analyzed in Figure 1 of “Allergic inflammatory memory in human respiratory epithelial progenitor cells“. Raw cell (columns) x gene (rows) matrix (@raw.data slot in Seurat) accompanied by the meta data table (@data.info slot in Seurat) […]...

scRAD supports a reproducibility-driven framework for assessing the expression differences in single-cell RNA-Seq (scRNA-Seq) data collected from multiple donors. scRAD provides tools for: Reproducible gene module analysis – identifying axes of variation common across donors Irreproducible Discovery Rate (IDR) analysis (Q. Li et al. 2011) for assessing signal reproducibility across multiple donors Other meta-analysis applicable to multi-donor studies […]...

In the Shalek Lab, we bring the same level of commitment and attention to detail we apply to our science to our cooking and cocktails. Here are recipes for 3 pepper-infused mezcals, ranked X, XX and XXX based on their mouth-numbing potential. We also make a 4th, X^X (recipe not shown), which is probably not […]...

Seq-Well is a portable, low-cost platform for single-cell RNA sequencing designed to be compatible with low-input, clinical biopsies. We have recently published a manuscript detailing the development and validation of the Seq-Well plaftorm. Here, we provide an in-depth protocol and videos describing how to perform Seq-Well experiments. We hope that you will use Seq-Well and apply it in […]...